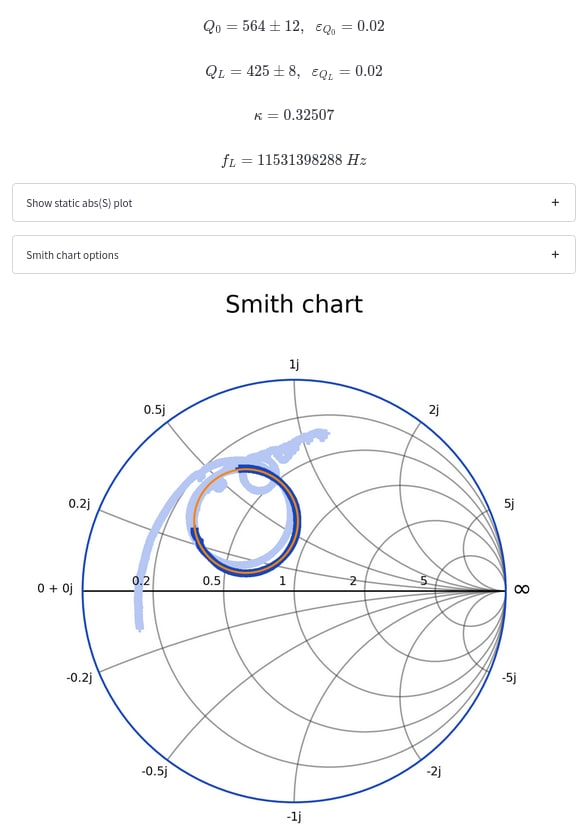
|
|
@@ -1,361 +1,135 @@
|
|
|
-import math
|
|
|
import streamlit as st
|
|
|
import matplotlib.pyplot as plt
|
|
|
+
|
|
|
import numpy as np
|
|
|
import sigfig
|
|
|
from streamlit_ace import st_ace
|
|
|
-from streamlit_echarts import st_echarts, JsCode
|
|
|
-
|
|
|
-
|
|
|
-# So that you can choose an interval of points on which we apply q-calc algorithm
|
|
|
-def plot_interact_abs_from_f(f, r, i, interval_range):
|
|
|
- if interval_range is None:
|
|
|
- interval_range = (0, 100)
|
|
|
-
|
|
|
- abs_S = list(abs(np.array(r) + 1j * np.array(i)))
|
|
|
- # echarts for datazoom https://discuss.streamlit.io/t/streamlit-echarts/3655
|
|
|
- # datazoom https://echarts.apache.org/examples/en/editor.html?c=line-draggable&lang=ts
|
|
|
- # axis pointer values https://echarts.apache.org/en/option.html#axisPointer
|
|
|
- options = {
|
|
|
- "xAxis": {
|
|
|
- "type": "category",
|
|
|
- "data": f,
|
|
|
- "name": "Hz",
|
|
|
- "nameTextStyle": {"fontSize": 16},
|
|
|
- "axisLabel": {"fontSize": 16},
|
|
|
- },
|
|
|
- "yAxis": {
|
|
|
- "type": "value",
|
|
|
- "name": "abs(S)",
|
|
|
- "nameTextStyle": {"fontSize": 16},
|
|
|
- "axisLabel": {"fontSize": 16},
|
|
|
- # "axisPointer": {
|
|
|
- # "type": 'cross',
|
|
|
- # "label": {
|
|
|
- # "show":"true",
|
|
|
- # "formatter": JsCode(
|
|
|
- # "function(info){console.log(info);return 'line ' ;};"
|
|
|
- # ).js_code
|
|
|
- # }
|
|
|
- # }
|
|
|
- },
|
|
|
- "series": [{"data": abs_S, "type": "line", "name": "abs(S)"}],
|
|
|
- "height": 300,
|
|
|
- "dataZoom": [{"type": "slider", "start": interval_range[0], "end": interval_range[1], "height": 100, "bottom": 10}],
|
|
|
- "tooltip": {
|
|
|
- "trigger": "axis",
|
|
|
- "axisPointer": {
|
|
|
- "type": 'cross',
|
|
|
- # "label": {
|
|
|
- # "show":"true",
|
|
|
- # "formatter": JsCode(
|
|
|
- # "function(info){console.log(info);return 'line ' ;};"
|
|
|
- # ).js_code
|
|
|
- # }
|
|
|
- }
|
|
|
- },
|
|
|
- "toolbox": {
|
|
|
- "feature": {
|
|
|
- # "dataView": { "show": "true", "readOnly": "true" },
|
|
|
- "restore": {"show": "true"},
|
|
|
- }
|
|
|
- },
|
|
|
- }
|
|
|
- # DataZoom event is not fired on new file upload. There are no default event to fix it.
|
|
|
- events = {
|
|
|
- "dataZoom": "function(params) { return ['dataZoom', params.start, params.end] }",
|
|
|
- "restore": "function() { return ['restore'] }",
|
|
|
- }
|
|
|
-
|
|
|
- # show echart with dataZoom and update intervals based on output
|
|
|
-
|
|
|
- get_event = st_echarts(
|
|
|
- options=options, events=events, height="500px", key="render_basic_bar_events"
|
|
|
- )
|
|
|
-
|
|
|
- if not get_event is None and get_event[0] == 'dataZoom':
|
|
|
- interval_range = get_event[1:]
|
|
|
-
|
|
|
- n = len(f)
|
|
|
- interval_start, interval_end = (
|
|
|
- int(n*interval_range[id]*0.01) for id in (0, 1))
|
|
|
- return interval_range, interval_start, interval_end
|
|
|
-
|
|
|
|
|
|
-
|
|
|
-def circle(ax, x, y, radius, color='#1946BA'):
|
|
|
- from matplotlib.patches import Ellipse
|
|
|
- drawn_circle = Ellipse((x, y), radius * 2, radius * 2, clip_on=True,
|
|
|
- zorder=2, linewidth=2, edgecolor=color, facecolor=(0, 0, 0, .0125))
|
|
|
- ax.add_artist(drawn_circle)
|
|
|
-
|
|
|
-
|
|
|
-def plot_smith(r, i, g, r_cut, i_cut, show_excluded):
|
|
|
+from .draw_smith_utils import draw_smith_circle, plot_abs_s_gridlines, plot_im_z_gridlines, plot_re_z_gridlines
|
|
|
+from .show_amplitude_echart import plot_interact_abs_from_f
|
|
|
+from .data_parsing_utils import parse_snp_header, read_data, count_columns, prepare_snp, unpack_data
|
|
|
+
|
|
|
+
|
|
|
+def plot_smith(r, i, g, r_cut, i_cut):
|
|
|
+ # maintaining state again (remember options for this session)
|
|
|
+ if 'smith_options' not in st.session_state:
|
|
|
+ st.session_state.smith_options = (True, True, False, False, False)
|
|
|
+ with st.expander("Smith chart options"):
|
|
|
+
|
|
|
+ smith_options_input = (st.checkbox(
|
|
|
+ "Show excluded points",
|
|
|
+ value=st.session_state.smith_options[0]),
|
|
|
+ st.checkbox("Show grid",
|
|
|
+ st.session_state.smith_options[1]),
|
|
|
+ st.checkbox(
|
|
|
+ "Show |S| gridlines",
|
|
|
+ value=st.session_state.smith_options[2],
|
|
|
+ ),
|
|
|
+ st.checkbox(
|
|
|
+ "Show Re(Z) gridlines",
|
|
|
+ value=st.session_state.smith_options[3],
|
|
|
+ ),
|
|
|
+ st.checkbox(
|
|
|
+ "Show Im(Z) gridlines",
|
|
|
+ value=st.session_state.smith_options[4],
|
|
|
+ ))
|
|
|
+ if st.session_state.smith_options != smith_options_input:
|
|
|
+ st.session_state.smith_options = smith_options_input
|
|
|
+ st.experimental_rerun()
|
|
|
+
|
|
|
+ (show_excluded_points, show_grid, show_Abs_S_gridlines,
|
|
|
+ show_Re_Z_gridlines, show_Im_Z_gridlines) = st.session_state.smith_options
|
|
|
fig = plt.figure(figsize=(10, 10))
|
|
|
ax = fig.add_subplot()
|
|
|
-
|
|
|
- # major_ticks = np.arange(-1.0, 1.1, 0.25)
|
|
|
+ ax.axis('equal')
|
|
|
minor_ticks = np.arange(-1.1, 1.1, 0.05)
|
|
|
- # ax.set_xticks(major_ticks)
|
|
|
ax.set_xticks(minor_ticks, minor=True)
|
|
|
- # ax.set_yticks(major_ticks)
|
|
|
ax.set_yticks(minor_ticks, minor=True)
|
|
|
ax.grid(which='major', color='grey', linewidth=1.5)
|
|
|
ax.grid(which='minor', color='grey', linewidth=0.5, linestyle=':')
|
|
|
- plt.xlabel('$Re(\Gamma)$', color='gray', fontsize=16, fontname="Cambria")
|
|
|
- plt.ylabel('$Im(\Gamma)$', color='gray', fontsize=16, fontname="Cambria")
|
|
|
+ plt.xlabel('$Re(S)$', color='gray', fontsize=16, fontname="Cambria")
|
|
|
+ plt.ylabel('$Im(S)$', color='gray', fontsize=16, fontname="Cambria")
|
|
|
plt.title('Smith chart', fontsize=24, fontname="Cambria")
|
|
|
|
|
|
# unit circle
|
|
|
- circle(ax, 0, 0, 1)
|
|
|
+ draw_smith_circle(ax, 0, 0, 1, '#1946BA')
|
|
|
+
|
|
|
+ if not show_grid:
|
|
|
+ ax.axis('off')
|
|
|
+
|
|
|
+ if show_Abs_S_gridlines:
|
|
|
+ # imshow is extremely slow, so draw it in place
|
|
|
+ plot_abs_s_gridlines(ax)
|
|
|
+
|
|
|
+ if show_Re_Z_gridlines:
|
|
|
+ plot_re_z_gridlines(ax)
|
|
|
+
|
|
|
+ if show_Im_Z_gridlines:
|
|
|
+ plot_im_z_gridlines(ax)
|
|
|
|
|
|
# input data points
|
|
|
- if show_excluded:
|
|
|
+ if show_excluded_points:
|
|
|
ax.plot(r, i, '+', ms=8, mew=2, color='#b6c7f4')
|
|
|
|
|
|
# choosen data points
|
|
|
ax.plot(r_cut, i_cut, '+', ms=8, mew=2, color='#1946BA')
|
|
|
|
|
|
- # circle approximation by calc
|
|
|
+ # S-circle approximation by calc
|
|
|
radius = abs(g[1] - g[0] / g[2]) / 2
|
|
|
x = ((g[1] + g[0] / g[2]) / 2).real
|
|
|
y = ((g[1] + g[0] / g[2]) / 2).imag
|
|
|
- circle(ax, x, y, radius, color='#FF8400')
|
|
|
+ draw_smith_circle(ax, x, y, radius, color='#FF8400')
|
|
|
|
|
|
- XLIM = [-1.1, 1.1]
|
|
|
- YLIM = [-1.1, 1.1]
|
|
|
+ XLIM = [-1.3, 1.3]
|
|
|
+ YLIM = [-1.3, 1.3]
|
|
|
ax.set_xlim(XLIM)
|
|
|
ax.set_ylim(YLIM)
|
|
|
- st.pyplot(fig)
|
|
|
+ try:
|
|
|
+ st.pyplot(fig)
|
|
|
+ except:
|
|
|
+ st.write('Unexpected plot error')
|
|
|
|
|
|
|
|
|
-# plot (abs(S))(f) chart with pyplot
|
|
|
+# plot abs(S) vs f chart with pyplot
|
|
|
def plot_abs_vs_f(f, r, i, fitted_mag_s):
|
|
|
fig = plt.figure(figsize=(10, 10))
|
|
|
- abs_S = list((r[n] ** 2 + i[n] ** 2)**0.5 for n in range(len(r)))
|
|
|
- xlim = [min(f) - abs(max(f) - min(f)) * 0.1,
|
|
|
- max(f) + abs(max(f) - min(f)) * 0.1]
|
|
|
- ylim = [min(abs_S) - abs(max(abs_S) - min(abs_S)) * 0.5,
|
|
|
- max(abs_S) + abs(max(abs_S) - min(abs_S)) * 0.5]
|
|
|
+ s = np.abs(np.array(r) + 1j * np.array(i))
|
|
|
+ if st.session_state.legendselection == '|S| (dB)':
|
|
|
+ m = np.min(np.where(s == 0, np.inf, s))
|
|
|
+ s = list(20 * np.where(s == 0, np.log10(m), np.log10(s)))
|
|
|
+ m = np.min(np.where(s == 0, np.inf, fitted_mag_s))
|
|
|
+ fitted_mag_s = list(
|
|
|
+ 20 * np.where(s == 0, np.log10(m), np.log10(fitted_mag_s)))
|
|
|
+ s = list(s)
|
|
|
+ min_f = min(f)
|
|
|
+ max_f = max(f)
|
|
|
+ xlim = [min_f - abs(max_f - min_f) * 0.1, max_f + abs(max_f - min_f) * 0.1]
|
|
|
+ min_s = min(s)
|
|
|
+ max_s = max(s)
|
|
|
+ ylim = [min_s - abs(max_s - min_s) * 0.5, max_s + abs(max_s - min_s) * 0.5]
|
|
|
ax = fig.add_subplot()
|
|
|
ax.set_xlim(xlim)
|
|
|
ax.set_ylim(ylim)
|
|
|
ax.grid(which='major', color='k', linewidth=1)
|
|
|
ax.grid(which='minor', color='grey', linestyle=':', linewidth=0.5)
|
|
|
plt.xlabel(r'$f,\; 1/c$', color='gray', fontsize=16, fontname="Cambria")
|
|
|
- plt.ylabel('$|S|$', color='gray', fontsize=16, fontname="Cambria")
|
|
|
- plt.title('Abs(S) vs frequency',
|
|
|
- fontsize=24, fontname="Cambria")
|
|
|
-
|
|
|
- ax.plot(f, abs_S, '+', ms=8, mew=2, color='#1946BA')
|
|
|
+ if st.session_state.legendselection == '|S| (dB)':
|
|
|
+ plt.ylabel('$|S|$ (dB)', color='gray', fontsize=16, fontname="Cambria")
|
|
|
+ plt.title('|S| (dB) vs frequency', fontsize=24, fontname="Cambria")
|
|
|
+ else:
|
|
|
+ plt.ylabel('$|S|$', color='gray', fontsize=16, fontname="Cambria")
|
|
|
+ plt.title('|S| vs frequency', fontsize=24, fontname="Cambria")
|
|
|
|
|
|
- ax.plot(f, fitted_mag_s, '-',ms=8, mew=8, color='#FF8400')
|
|
|
+ ax.plot(f, s, '+', ms=8, mew=2, color='#1946BA')
|
|
|
|
|
|
- # radius = abs(g[1] - g[0] / g[2]) / 2
|
|
|
- # x = ((g[1] + g[0] / g[2]) / 2).real
|
|
|
- # y = ((g[1] + g[0] / g[2]) / 2).imag
|
|
|
- st.pyplot(fig)
|
|
|
+ ax.plot(f, fitted_mag_s, '-', linewidth=3, color='#FF8400')
|
|
|
|
|
|
+ try:
|
|
|
+ st.pyplot(fig)
|
|
|
+ except:
|
|
|
+ st.write('Unexpected plot error')
|
|
|
|
|
|
def run(calc_function):
|
|
|
-
|
|
|
- def is_float(element) -> bool:
|
|
|
- try:
|
|
|
- float(element)
|
|
|
- val = float(element)
|
|
|
- if math.isnan(val) or math.isinf(val):
|
|
|
- raise ValueError
|
|
|
- return True
|
|
|
- except ValueError:
|
|
|
- return False
|
|
|
-
|
|
|
- # to utf-8
|
|
|
- def read_data(data):
|
|
|
- for x in range(len(data)):
|
|
|
- if type(data[x]) == bytes:
|
|
|
- try:
|
|
|
- data[x] = data[x].decode('utf-8-sig', 'ignore')
|
|
|
- except:
|
|
|
- return 'Not an utf-8-sig line №: ' + str(x)
|
|
|
- return 'data read, but not parsed'
|
|
|
-
|
|
|
- # for Touchstone .snp format
|
|
|
- def parse_heading(data):
|
|
|
- nonlocal data_format_snp
|
|
|
- if data_format_snp:
|
|
|
- for x in range(len(data)):
|
|
|
- if data[x][0] == '#':
|
|
|
- line = data[x].split()
|
|
|
- if len(line) == 6:
|
|
|
- repr_map = {"RI": 0, "MA": 1, "DB": 2}
|
|
|
- para_map = {"S": 0, "Z": 1}
|
|
|
- hz_map = {"GHz": 10**9, "MHz": 10 **6, "KHz": 10**3, "Hz": 1}
|
|
|
- hz, measurement_parameter, data_representation, _r, ref_resistance = line[1:]
|
|
|
- try:
|
|
|
- return hz_map[hz], para_map[measurement_parameter], repr_map[data_representation], int(ref_resistance)
|
|
|
- except:
|
|
|
- break
|
|
|
- break
|
|
|
- return 1, 0, 0, 50
|
|
|
-
|
|
|
- # check if line has comments
|
|
|
- # first is a comment line according to .snp documentation,
|
|
|
- # others detects comments in various languages
|
|
|
- def check_line_comments(line):
|
|
|
- if len(line) < 2 or line[0] == '!' or line[0] == '#' or line[0] == '%' or line[0] == '/':
|
|
|
- return None
|
|
|
- else:
|
|
|
- # generally we expect these chars as separators
|
|
|
- line = line.replace(';', ' ').replace(',', ' ')
|
|
|
- if '!' in line:
|
|
|
- line = line[:line.find('!')]
|
|
|
- return line
|
|
|
-
|
|
|
- # unpack a few first lines of the file to get number of ports
|
|
|
- def count_columns(data):
|
|
|
- return_status = 'data parsed'
|
|
|
- column_count = 0
|
|
|
- for x in range(len(data)):
|
|
|
- line = check_line_comments(data[x])
|
|
|
- if line is None:
|
|
|
- continue
|
|
|
- line = line.split()
|
|
|
- # always at least 3 values for single data point
|
|
|
- if len(line) < 3:
|
|
|
- return_status = 'Can\'t parse line № ' + \
|
|
|
- str(x) + ',\n not enough arguments (less than 3)'
|
|
|
- break
|
|
|
- column_count = len(line)
|
|
|
- break
|
|
|
- return column_count, return_status
|
|
|
-
|
|
|
- def prepare_snp(data, number):
|
|
|
- prepared_data = []
|
|
|
- return_status = 'data read, but not parsed'
|
|
|
- for x in range(len(data)):
|
|
|
- line = check_line_comments(data[x])
|
|
|
- if line is None:
|
|
|
- continue
|
|
|
-
|
|
|
- splitted_line = line.split()
|
|
|
- if number * 2 + 1 == len(splitted_line):
|
|
|
- prepared_data.append(line)
|
|
|
- elif number * 2 == len(splitted_line):
|
|
|
- prepared_data[-1] += line
|
|
|
- else:
|
|
|
- return_status = "Parsing error for .snp format on line №" + str(x)
|
|
|
-
|
|
|
- return prepared_data, return_status
|
|
|
-
|
|
|
- def unpack_data(data, first_column, column_count, ref_resistance, ace_preview_markers):
|
|
|
- nonlocal select_measurement_parameter
|
|
|
- nonlocal select_data_representation
|
|
|
- f, r, i = [], [], []
|
|
|
- return_status = 'data parsed'
|
|
|
- for x in range(len(data)):
|
|
|
- line = check_line_comments(data[x])
|
|
|
- if line is None:
|
|
|
- continue
|
|
|
-
|
|
|
- line = line.split()
|
|
|
-
|
|
|
- if column_count != len(line):
|
|
|
- return_status = "Wrong number of parameters on line № " + str(x)
|
|
|
- break
|
|
|
-
|
|
|
- # 1: process according to data_placement
|
|
|
- a, b, c = None, None, None
|
|
|
- try:
|
|
|
- a = line[0]
|
|
|
- b = line[first_column]
|
|
|
- c = line[first_column+1]
|
|
|
- except:
|
|
|
- return_status = 'Can\'t parse line №: ' + \
|
|
|
- str(x) + ',\n not enough arguments'
|
|
|
- break
|
|
|
- if not ((is_float(a)) or (is_float(b)) or (is_float(c))):
|
|
|
- return_status = 'Wrong data type, expected number. Error on line: ' + \
|
|
|
- str(x)
|
|
|
- break
|
|
|
-
|
|
|
- # mark as processed
|
|
|
- for y in (a,b,c):
|
|
|
- ace_preview_markers.append(
|
|
|
- {"startRow": x,"startCol": 0,
|
|
|
- "endRow": x,"endCol": data[x].find(y)+len(y),
|
|
|
- "className": "ace_stack","type": "text"})
|
|
|
-
|
|
|
- a, b, c = (float(x) for x in (a, b, c))
|
|
|
- f.append(a) # frequency
|
|
|
-
|
|
|
- # 2: process according to data_representation
|
|
|
- if select_data_representation == 'Frequency, real, imaginary':
|
|
|
- # std format
|
|
|
- r.append(b) # Re
|
|
|
- i.append(c) # Im
|
|
|
- elif select_data_representation == 'Frequency, magnitude, angle':
|
|
|
- r.append(b*np.cos(np.deg2rad(c)))
|
|
|
- i.append(b*np.sin(np.deg2rad(c)))
|
|
|
- elif select_data_representation == 'Frequency, db, angle':
|
|
|
- b = 10**(b/20)
|
|
|
- r.append(b*np.cos(np.deg2rad(c)))
|
|
|
- i.append(b*np.sin(np.deg2rad(c)))
|
|
|
- else:
|
|
|
- return_status = 'Wrong data format'
|
|
|
- break
|
|
|
-
|
|
|
- # 3: process according to measurement_parameter
|
|
|
- if select_measurement_parameter == 'Z':
|
|
|
- # normalization
|
|
|
- r[-1] = r[-1]/ref_resistance
|
|
|
- i[-1] = i[-1]/ref_resistance
|
|
|
-
|
|
|
- # translate to S
|
|
|
- try:
|
|
|
- # center_x + 1j*center_y, radius
|
|
|
- p1, r1 = r[-1] / (1 + r[-1]) + 0j, 1 / (1 + r[-1]) #real
|
|
|
- p2, r2 = 1 + 1j * (1 / i[-1]), 1 / i[-1] #imag
|
|
|
-
|
|
|
- d = abs(p2-p1)
|
|
|
- q = (r1**2 - r2**2 + d**2) / (2 * d)
|
|
|
-
|
|
|
- h = (r1**2 - q**2)**0.5
|
|
|
-
|
|
|
- p = p1 + q * (p2 - p1) / d
|
|
|
-
|
|
|
- intersect = [
|
|
|
- (p.real + h * (p2.imag - p1.imag) / d,
|
|
|
- p.imag - h * (p2.real - p1.real) / d),
|
|
|
- (p.real - h * (p2.imag - p1.imag) / d,
|
|
|
- p.imag + h * (p2.real - p1.real) / d)]
|
|
|
-
|
|
|
- intersect = [x+1j*y for x,y in intersect]
|
|
|
- intersect_shift = [p-(1+0j) for p in intersect]
|
|
|
- intersect_shift = abs(np.array(intersect_shift))
|
|
|
- p=intersect[0]
|
|
|
- if intersect_shift[0]<intersect_shift[1]:
|
|
|
- p=intersect[1]
|
|
|
- r[-1] = p.real
|
|
|
- i[-1] = p.imag
|
|
|
- except:
|
|
|
- r.pop()
|
|
|
- i.pop()
|
|
|
- f.pop()
|
|
|
-
|
|
|
- if return_status == 'data parsed':
|
|
|
- if len(f) < 3 or len(f) != len(r) or len(f) != len(i):
|
|
|
- return_status = 'Choosen data range is too small, add more points'
|
|
|
- elif max(abs(np.array(r)+ 1j* np.array(i))) > 2:
|
|
|
- return_status = 'Your data points have an abnormality:\
|
|
|
- they are too far outside the unit cirlce.\
|
|
|
- Make sure the format is correct'
|
|
|
-
|
|
|
- return f, r, i, return_status
|
|
|
-
|
|
|
- # make accessible a specific range of numerical data choosen with interactive plot
|
|
|
- # percent, line id, line id
|
|
|
- interval_range, interval_start, interval_end = None, None, None
|
|
|
-
|
|
|
# info
|
|
|
with st.expander("Info"):
|
|
|
# streamlit.markdown does not support footnotes
|
|
|
@@ -366,33 +140,36 @@ def run(calc_function):
|
|
|
st.write('Wrong start directory, see readme')
|
|
|
|
|
|
# file upload button
|
|
|
- uploaded_file = st.file_uploader('Upload a file from your vector analizer. \
|
|
|
- Make sure the file format is .snp or it has a similar inner structure.' )
|
|
|
+ uploaded_file = st.file_uploader(
|
|
|
+ 'Upload a file from your vector analizer. \
|
|
|
+ Make sure the file format is .snp or it has a similar inner structure.'
|
|
|
+ )
|
|
|
|
|
|
# check .snp
|
|
|
- data_format_snp = False
|
|
|
+ is_data_format_snp = False
|
|
|
data_format_snp_number = 0
|
|
|
if uploaded_file is None:
|
|
|
st.write("DEMO: ")
|
|
|
# display DEMO
|
|
|
- data_format_snp = True
|
|
|
+ is_data_format_snp = True
|
|
|
try:
|
|
|
with open('./resource/data/8_default_demo.s1p') as f:
|
|
|
data = f.readlines()
|
|
|
except:
|
|
|
# 'streamlit run' call in the wrong directory. Display smaller demo:
|
|
|
- data =['# Hz S MA R 50\n\
|
|
|
+ data = [
|
|
|
+ '# Hz S MA R 50\n\
|
|
|
11415403125 0.37010744 92.47802\n\
|
|
|
11416090625 0.33831283 92.906929\n\
|
|
|
- 11416778125 0.3069371 94.03318' ]
|
|
|
+ 11416778125 0.3069371 94.03318'
|
|
|
+ ]
|
|
|
else:
|
|
|
data = uploaded_file.readlines()
|
|
|
- if uploaded_file.name[-4:-2]=='.s' and uploaded_file.name[-1]== 'p':
|
|
|
- data_format_snp = True
|
|
|
+ if uploaded_file.name[-4:-2] == '.s' and uploaded_file.name[-1] == 'p':
|
|
|
+ is_data_format_snp = True
|
|
|
data_format_snp_number = int(uploaded_file.name[-2])
|
|
|
|
|
|
validator_status = '...'
|
|
|
- ace_preview_markers = []
|
|
|
column_count = 0
|
|
|
|
|
|
# data loaded
|
|
|
@@ -401,34 +178,51 @@ def run(calc_function):
|
|
|
|
|
|
validator_status = read_data(data)
|
|
|
if validator_status == 'data read, but not parsed':
|
|
|
- hz, select_measurement_parameter, select_data_representation, input_ref_resistance = parse_heading(data)
|
|
|
+ hz, select_measurement_parameter, select_data_representation, input_ref_resistance = parse_snp_header(
|
|
|
+ data, is_data_format_snp)
|
|
|
|
|
|
- col1, col2 = st.columns([1,2])
|
|
|
+ col1, col2 = st.columns([1, 2])
|
|
|
|
|
|
ace_text_value = ''.join(data).strip()
|
|
|
with col1.expander("Processing options"):
|
|
|
- select_measurement_parameter = st.selectbox('Measurement parameter',
|
|
|
- ['S', 'Z'],
|
|
|
- select_measurement_parameter)
|
|
|
- select_data_representation = st.selectbox('Data representation',
|
|
|
- ['Frequency, real, imaginary',
|
|
|
- 'Frequency, magnitude, angle',
|
|
|
- 'Frequency, db, angle'],
|
|
|
- select_data_representation)
|
|
|
- if select_measurement_parameter=='Z':
|
|
|
+ select_measurement_parameter = st.selectbox(
|
|
|
+ 'Measurement parameter', ['S', 'Z'],
|
|
|
+ select_measurement_parameter)
|
|
|
+ select_data_representation = st.selectbox(
|
|
|
+ 'Data representation', [
|
|
|
+ 'Frequency, real, imaginary',
|
|
|
+ 'Frequency, magnitude, angle', 'Frequency, db, angle'
|
|
|
+ ], select_data_representation)
|
|
|
+ if select_measurement_parameter == 'Z':
|
|
|
input_ref_resistance = st.number_input(
|
|
|
- "Reference resistance:", min_value=0, value=input_ref_resistance)
|
|
|
- input_start_line = int(st.number_input(
|
|
|
- "First line for processing:", min_value=1, max_value=len(data)))
|
|
|
- input_end_line = int(st.number_input(
|
|
|
- "Last line for processing:", min_value=1, max_value=len(data), value=len(data)))
|
|
|
- data = data[input_start_line-1:input_end_line]
|
|
|
-
|
|
|
+ "Reference resistance:",
|
|
|
+ min_value=0,
|
|
|
+ value=input_ref_resistance)
|
|
|
+ if not is_data_format_snp:
|
|
|
+ input_hz = st.selectbox('Unit of frequency',
|
|
|
+ ['Hz', 'KHz', 'MHz', 'GHz'], 0)
|
|
|
+ hz_map = {
|
|
|
+ "ghz": 10**9,
|
|
|
+ "mhz": 10**6,
|
|
|
+ "khz": 10**3,
|
|
|
+ "hz": 1
|
|
|
+ }
|
|
|
+ hz = hz_map[input_hz.lower()]
|
|
|
+ input_start_line = int(
|
|
|
+ st.number_input("First line for processing:",
|
|
|
+ min_value=1,
|
|
|
+ max_value=len(data)))
|
|
|
+ input_end_line = int(
|
|
|
+ st.number_input("Last line for processing:",
|
|
|
+ min_value=1,
|
|
|
+ max_value=len(data),
|
|
|
+ value=len(data)))
|
|
|
+ if input_end_line < input_start_line:
|
|
|
+ input_end_line=input_start_line
|
|
|
+ data = data[input_start_line - 1:input_end_line]
|
|
|
|
|
|
# Ace editor to show choosen data columns and rows
|
|
|
with col2.expander("File preview"):
|
|
|
- # st.button(copy selection)
|
|
|
-
|
|
|
# So little 'official' functionality in libs and lack of documentation
|
|
|
# therefore beware: css hacks
|
|
|
|
|
|
@@ -445,20 +239,22 @@ def run(calc_function):
|
|
|
# color: rgb(49, 51, 63);
|
|
|
# }</style>''', unsafe_allow_html=True)
|
|
|
|
|
|
- # markdown injection does not seems to work, since ace is in a different .html accessible via iframe
|
|
|
+ # markdown injection does not seems to work,
|
|
|
+ # since ace is in a different .html accessible via iframe
|
|
|
|
|
|
# markers format:
|
|
|
#[{"startRow": 2,"startCol": 0,"endRow": 2,"endCol": 3,"className": "ace_error-marker","type": "text"}]
|
|
|
|
|
|
- # add marking for choosen data lines TODO
|
|
|
- ace_preview_markers.append({
|
|
|
+ # add marking for choosen data lines?
|
|
|
+ # todo or not todo?
|
|
|
+ ace_preview_markers =[{
|
|
|
"startRow": input_start_line - 1,
|
|
|
"startCol": 0,
|
|
|
"endRow": input_end_line,
|
|
|
"endCol": 0,
|
|
|
"className": "ace_highlight-marker",
|
|
|
"type": "text"
|
|
|
- })
|
|
|
+ }]
|
|
|
|
|
|
st_ace(value=ace_text_value,
|
|
|
readonly=True,
|
|
|
@@ -467,8 +263,9 @@ def run(calc_function):
|
|
|
markers=ace_preview_markers,
|
|
|
height="300px")
|
|
|
|
|
|
- if data_format_snp and data_format_snp_number >= 3:
|
|
|
- data, validator_status = prepare_snp(data, data_format_snp_number)
|
|
|
+ if is_data_format_snp and data_format_snp_number >= 3:
|
|
|
+ data, validator_status = prepare_snp(data,
|
|
|
+ data_format_snp_number)
|
|
|
|
|
|
if validator_status == "data read, but not parsed":
|
|
|
column_count, validator_status = count_columns(data)
|
|
|
@@ -479,40 +276,47 @@ def run(calc_function):
|
|
|
if column_count > 3:
|
|
|
pair_count = (column_count - 1) // 2
|
|
|
input_ports_pair = st.number_input(
|
|
|
- "Choosen pair of ports with network parameters:",
|
|
|
+ "Choose pair of ports with network parameters:",
|
|
|
min_value=1,
|
|
|
max_value=pair_count,
|
|
|
value=1)
|
|
|
input_ports_pair_id = input_ports_pair - 1
|
|
|
ports_count = round(pair_count**0.5)
|
|
|
- st.write(select_measurement_parameter +
|
|
|
+ st.write('Choosen ports: ' + select_measurement_parameter +
|
|
|
str(input_ports_pair_id // ports_count + 1) +
|
|
|
str(input_ports_pair_id % ports_count + 1))
|
|
|
f, r, i, validator_status = unpack_data(data,
|
|
|
(input_ports_pair - 1) * 2 + 1,
|
|
|
column_count,
|
|
|
input_ref_resistance,
|
|
|
- ace_preview_markers)
|
|
|
+ select_measurement_parameter,
|
|
|
+ select_data_representation)
|
|
|
f = [x * hz for x in f] # to hz
|
|
|
|
|
|
st.write("Use range slider to choose best suitable data interval")
|
|
|
- interval_range, interval_start, interval_end = plot_interact_abs_from_f(f, r, i, interval_range)
|
|
|
-
|
|
|
- f_cut, r_cut, i_cut = [], [], []
|
|
|
- if validator_status == "data parsed":
|
|
|
- f_cut, r_cut, i_cut = (x[interval_start:interval_end]
|
|
|
- for x in (f, r, i))
|
|
|
|
|
|
- with st.expander("Selected data interval as .s1p"):
|
|
|
- st_ace(value="# Hz S RI R 50\n" +
|
|
|
- ''.join(f'{f_cut[x]} {r_cut[x]} {i_cut[x]}\n' for x in range(len(f_cut))),
|
|
|
+ if len(f)==0:
|
|
|
+ validator_status = 'data unpacking error: empty data'
|
|
|
+ else:
|
|
|
+ # make accessible a specific range of numerical data choosen with interactive plot
|
|
|
+ # line id, line id
|
|
|
+ interval_start, interval_end = plot_interact_abs_from_f(f,r,i)
|
|
|
+
|
|
|
+ f_cut, r_cut, i_cut = [], [], []
|
|
|
+ if validator_status == "data parsed":
|
|
|
+ f_cut, r_cut, i_cut = (x[interval_start:interval_end]
|
|
|
+ for x in (f, r, i))
|
|
|
+ with st.expander("Selected data interval as .s1p"):
|
|
|
+ st_ace(value="# Hz S RI R 50\n" +
|
|
|
+ ''.join(f'{f_cut[x]} {r_cut[x]} {i_cut[x]}\n'
|
|
|
+ for x in range(len(f_cut))),
|
|
|
readonly=True,
|
|
|
auto_update=True,
|
|
|
placeholder="Selection is empty",
|
|
|
height="150px")
|
|
|
-
|
|
|
- if len(f_cut) < 3:
|
|
|
- validator_status = "Choosen interval is too small, add more points"
|
|
|
+
|
|
|
+ if len(f_cut) < 3:
|
|
|
+ validator_status = "Choosen interval is too small, add more points"
|
|
|
|
|
|
st.write("Status: " + validator_status)
|
|
|
|
|
|
@@ -520,7 +324,7 @@ def run(calc_function):
|
|
|
col1, col2 = st.columns(2)
|
|
|
|
|
|
check_coupling_loss = col1.checkbox(
|
|
|
- 'Apply correction for coupling loss')
|
|
|
+ 'Apply correction for coupling losses', value = False)
|
|
|
|
|
|
if check_coupling_loss:
|
|
|
col1.write("Option: Lossy coupling")
|
|
|
@@ -528,41 +332,51 @@ def run(calc_function):
|
|
|
col1.write("Option: Cable attenuation")
|
|
|
|
|
|
select_autoformat = col2.checkbox("Autoformat output", value=True)
|
|
|
- precision = None
|
|
|
+ precision = '0.0f'
|
|
|
if not select_autoformat:
|
|
|
- precision = col2.slider("Precision", min_value=0, max_value=7, value = 4)
|
|
|
- precision = '0.'+str(precision)+'f'
|
|
|
+ precision = col2.slider("Precision",
|
|
|
+ min_value=0,
|
|
|
+ max_value=7,
|
|
|
+ value=4)
|
|
|
+ precision = '0.' + str(precision) + 'f'
|
|
|
|
|
|
- Q0, sigmaQ0, QL, sigmaQL, circle_params, fl, fitted_mag_s = calc_function(
|
|
|
+ Q0, sigmaQ0, QL, sigmaQL, k, ks, circle_params, fl, fitted_mag_s = calc_function(
|
|
|
f_cut, r_cut, i_cut, check_coupling_loss)
|
|
|
|
|
|
if Q0 <= 0 or QL <= 0:
|
|
|
st.write("Negative Q detected, fitting may be inaccurate!")
|
|
|
|
|
|
- if select_autoformat:
|
|
|
- st.latex(
|
|
|
- r'Q_0 =' +
|
|
|
- f'{sigfig.round(Q0, uncertainty=sigmaQ0, style="PDG")}, '
|
|
|
- + r'\;\;\varepsilon_{Q_0} =' +
|
|
|
- f'{sigfig.round(sigmaQ0 / Q0, sigfigs=1, style="PDG")}')
|
|
|
- st.latex(
|
|
|
- r'Q_L =' +
|
|
|
- f'{sigfig.round(QL, uncertainty=sigmaQL, style="PDG")}, '
|
|
|
- + r'\;\;\varepsilon_{Q_L} =' +
|
|
|
- f'{sigfig.round(sigmaQL / QL, sigfigs=1, style="PDG")}')
|
|
|
- else:
|
|
|
- st.latex(
|
|
|
- r'Q_0 =' +
|
|
|
- f'{format(Q0, precision)} \pm ' + f'{format(sigmaQ0, precision)}, '
|
|
|
- + r'\;\;\varepsilon_{Q_0} =' +
|
|
|
- f'{format(sigmaQ0 / Q0, precision)}')
|
|
|
- st.latex(
|
|
|
- r'Q_L =' +
|
|
|
- f'{format(QL, precision)} \pm ' + f'{format(sigmaQL, precision)}, '
|
|
|
- + r'\;\;\varepsilon_{Q_L} =' +
|
|
|
- f'{format(sigmaQL / QL, precision)}')
|
|
|
- st.latex(r'f_L ='+f'{fl}'+'Hz')
|
|
|
+ def show_result_in_latex(name, value, uncertainty=None):
|
|
|
+ nonlocal select_autoformat
|
|
|
+ if uncertainty is not None:
|
|
|
+ if select_autoformat:
|
|
|
+ st.latex(
|
|
|
+ name + ' =' +
|
|
|
+ f'{sigfig.round(value, uncertainty=uncertainty, style="PDG")}, '
|
|
|
+ + r'\;\;\varepsilon_{' + name + '} =' +
|
|
|
+ f'{sigfig.round(uncertainty / value, sigfigs=1, style="PDG")}'
|
|
|
+ )
|
|
|
+ else:
|
|
|
+ st.latex(name + ' =' + f'{format(value, precision)} \pm ' +
|
|
|
+ f'{format(uncertainty, precision)}, ' +
|
|
|
+ r'\;\;\varepsilon_{' + name + '} =' +
|
|
|
+ f'{format(uncertainty / value, precision)}')
|
|
|
+ else:
|
|
|
+ if select_autoformat:
|
|
|
+ st.latex(name + ' =' +
|
|
|
+ f'{sigfig.round(value, sigfigs=5, style="PDG")}')
|
|
|
+ else:
|
|
|
+ st.latex(name + ' =' + f'{format(value, precision)}')
|
|
|
+
|
|
|
+ show_result_in_latex('Q_0', Q0, sigmaQ0)
|
|
|
+ show_result_in_latex('Q_L', QL, sigmaQL)
|
|
|
+ show_result_in_latex(r'\kappa', k)
|
|
|
+ if check_coupling_loss:
|
|
|
+ show_result_in_latex(r'\kappa_s', ks)
|
|
|
+
|
|
|
+ st.latex('f_L =' + f'{format(fl, precision)}' + r'\text{ }Hz')
|
|
|
+
|
|
|
with st.expander("Show static abs(S) plot"):
|
|
|
plot_abs_vs_f(f_cut, r_cut, i_cut, fitted_mag_s)
|
|
|
|
|
|
- plot_smith(r, i, circle_params, r_cut, i_cut, st.checkbox("Show excluded points", value=True))
|
|
|
+ plot_smith(r, i, circle_params, r_cut, i_cut)
|